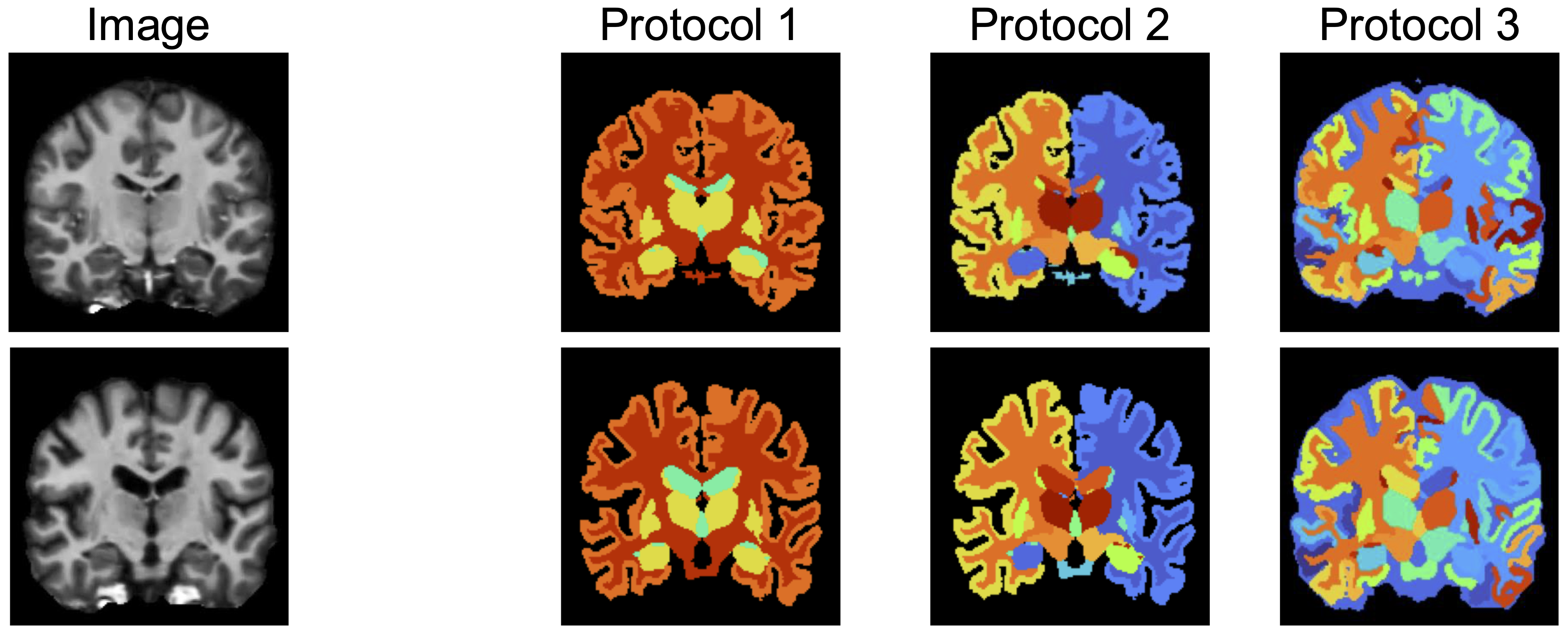
Overview
We introduce a new challenging task: for unseen biomedical domains, generate label maps from multiple protocols that are consistent across subjects.
Pancakes addresses it through a protocol-sampling mechanism and outperforms foundation segmentation models.